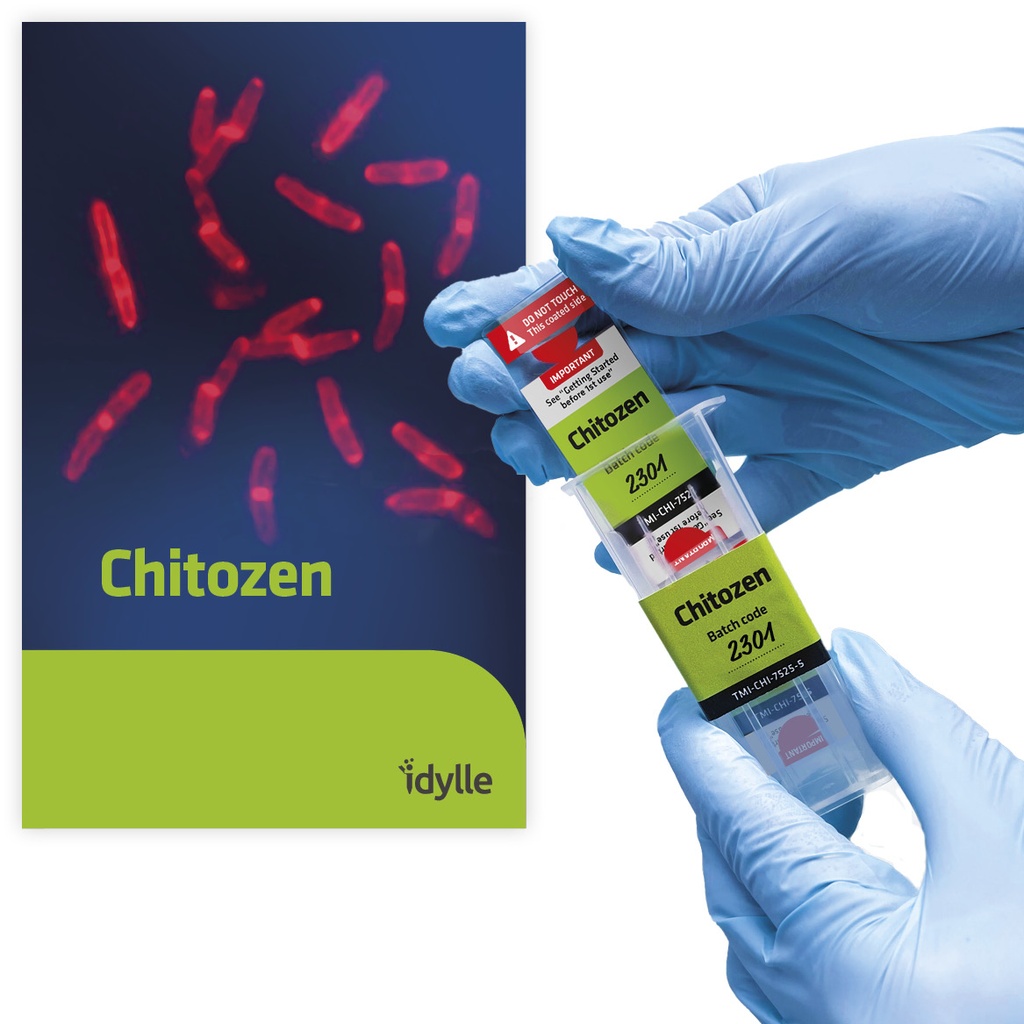
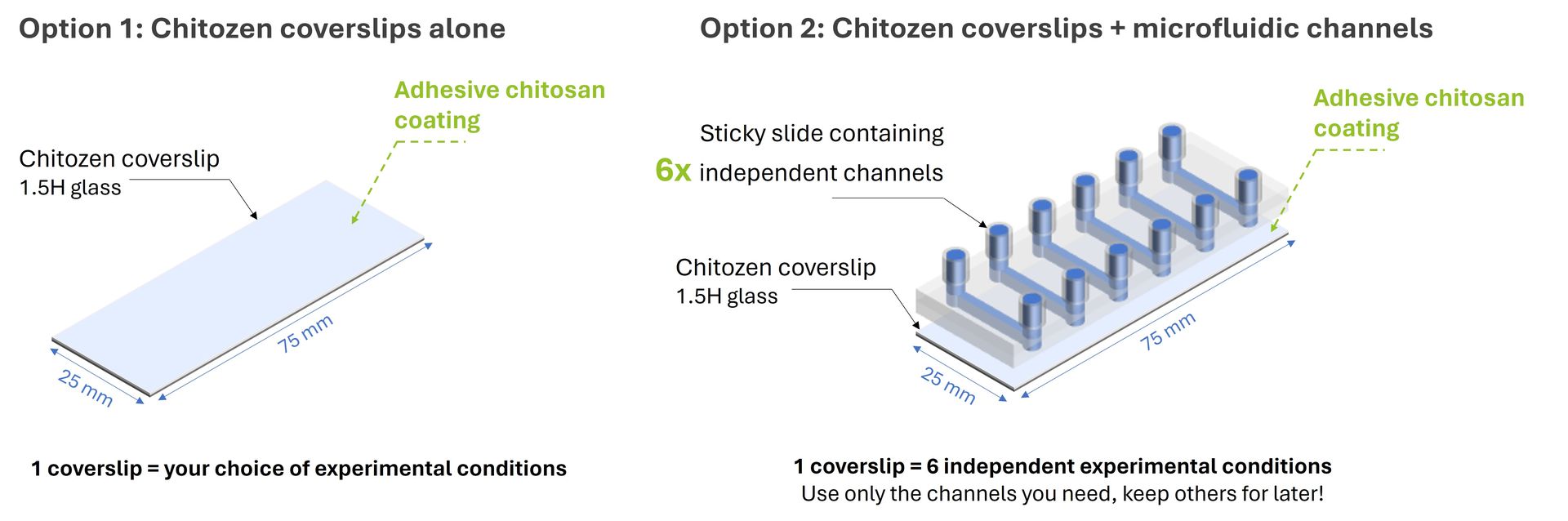
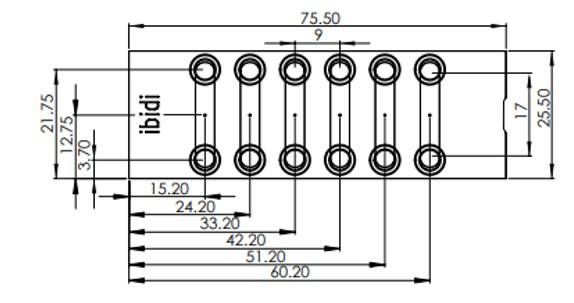
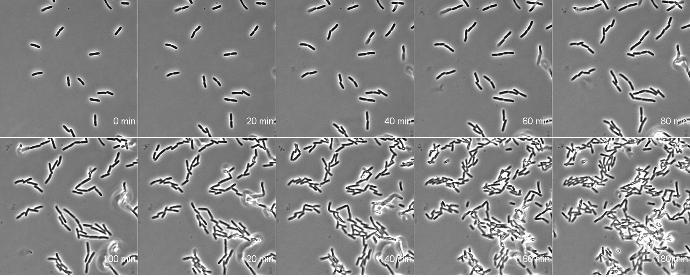
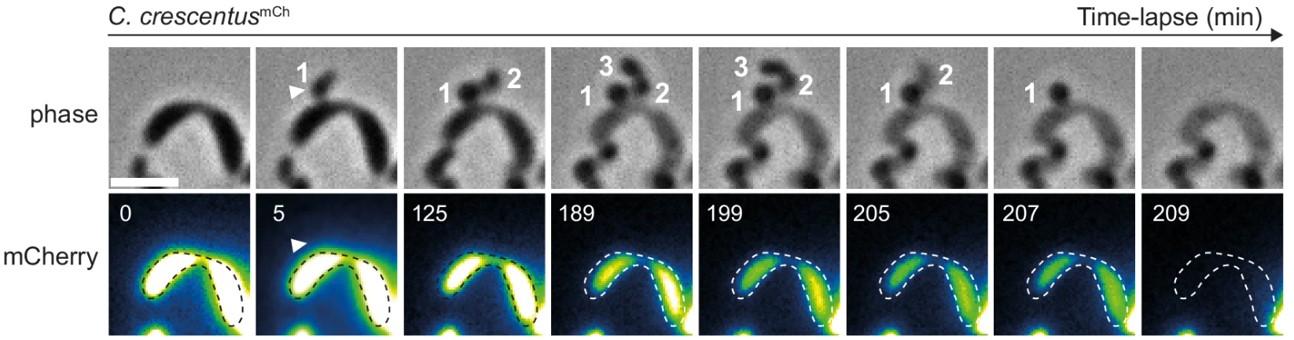
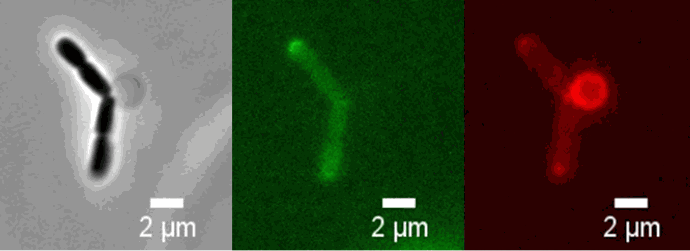
Chitozen - Adhesive coverslips for bacteria imaging
Chitosan-coated coverslips for live bacteria imaging
For any other kit format request, please contact us
A technology designed by Tâm Mignot, Olivier Theodoly, Amandine Desorme, Guillaume Sudre and Laurent David.
Everspark technology has been intially developed by Karine Monier, Arnaud Favier and Christophe Place and and published in Scientific Reports:
Provost, A., Rousset, C., Bourdon, L. et al. Innovative particle standards and long-lived imaging for 2D and 3D dSTORM. Sci Rep 9, 17967 (2019). https://doi.org/10.1038/s41598-019-53528-0
Publications:
Nanoscale engagement and clusterization of Programmed death ligand 1 (PD-L1) in the membrane lipid rafts of Non-Small Cell Lung Cancer cells
Martina Ruglioni, Simone Civita, Tiziano Salvadori, Sofia Cristiani, Vittoria Carnicelli, Serena Barachini, Iacopo Petrini, Irene Nepita, Marco Castello, Alberto Diaspro, Paolo Bianchini, Barbara Storti, Ranieri Bizzarri, Stefano Fogli and Romano Danesi bioRxiv 2022.08.09.503318; doi: https://doi.org/10.1101/2022.08.09.503318
HIV-1 diverts actin debranching mechanisms for particle assembly and release in CD4 T lymphocytes
Rayane Dibsy, Erwan Bremaud, Johnson Mak, Cyril Favard, Delphine Muriaux
BioRxiv, December 16, 2022. doi. 10.1101/2022.12.15.520580
Fluorescent Polymer-AS1411-Aptamer Probe for dSTORM Super-Resolution Imaging of Endogenous Nucleolin
Fabre L, Rousset C, Monier K, Da Cruz-Boisson F, Bouvet P, Charreyre MT, Delair T, Fleury E, Favier A. Biomacromolecules. 2022 May 12. doi: 10.1021/acs.biomac.1c01706. PMID: 35549176
Comparative analysis of ChAdOx1 nCoV-19 and Ad26.COV2.S SARS-CoV-2 vector vaccines.
Michalik S, Siegerist F, Palankar R, Franzke K, Schindler M, Reder A, Seifert U, Cammann C, Wesche J, Steil L, Hentschker C, Gesell-Salazar M, Reisinger E, Beer M, Endlich N, Greinacher A, Völker U. Haematologica. 2022 Apr 1;107(4):947-957. doi: 10.3324/haematol.2021.280154. PMID: 35045692
Metabolic biorthogonal labeling and dSTORM imaging of peptidoglycan synthesis in Streptococcus pneumoniae
Jennyfer Trouve, Oleksandr Glushonkov and Cecile Morlot
Star Protocols, December 13, 2021. doi: 10.1016/j.xpro.2021.101006
Insights in ChAdOx1 nCoV-19 vaccine-induced immune thrombotic thrombocytopenia.
Greinacher A, Selleng K, Palankar R, Wesche J, Handtke S, Wolff M, Aurich K, Lalk M, Methling K, Völker U, Hentschker C, Michalik S, Steil L, Reder A, Schönborn L, Beer M, Franzke K, Büttner A, Fehse B, Stavrou EX, Rangaswamy C, Mailer RK, Englert H, Frye M, Thiele T, Kochanek S, Krutzke L, Siegerist F, Endlich N, Warkentin TE, Renné T.
Blood. 2021 Dec 2;138(22):2256-2268. doi: 10.1182/blood.2021013231. PMID: 34587242
Superresolution Microscopy of Drosophila Indirect Flight Muscle Sarcomeres.
Szikora S, Novák T, Gajdos T, Erdélyi M, Mihály J. Bio Protoc. 2020 Jun 20;10(12):e3654. doi: 10.21769/BioProtoc.3654. eCollection 2020 Jun 20. PMID: 33659324