Elf lab in Uppsala (Sweden) designed a microfluidic device to capture individual bacterial cells from complex samples and to grow them with and without a candidate antibiotic. The average growth rates, with and without candidate antibiotic, are calculated in real time. The bacteria are considered susceptible if its growth is duly inhibited. This enables an incredibly fast (less than 30 mins) Antibiotic Susceptibility Test (AST). In general, this tool is very efficient to enable evidence-based prescription of antibiotics and help reduce the spread of antimicrobial resistance (AMR).
Publication:
Antibiotic susceptibility testing in less than 30min using direct single-cell imaging Proc. Natl. Acad. Sci., 114 (34) (2017), pp. 9170-9175
#PointOfCare #UTI #AST #Antibiotic #Resistance #Microfluidic

A blog post by Ilaria Iuliani
I am a physicist interested in biology. I have always been fascinated by the study of the mechanisms behind the life of microorganisms and how their understanding leads to great discoveries in different fields.
During my PhD, I developed a microfluidic setup to study the replication-division control in E.coli. This project has fueled my passion for innovative research tools, and I am now thrilled to share my favourite tools with you, thanks to the Idylle blog!
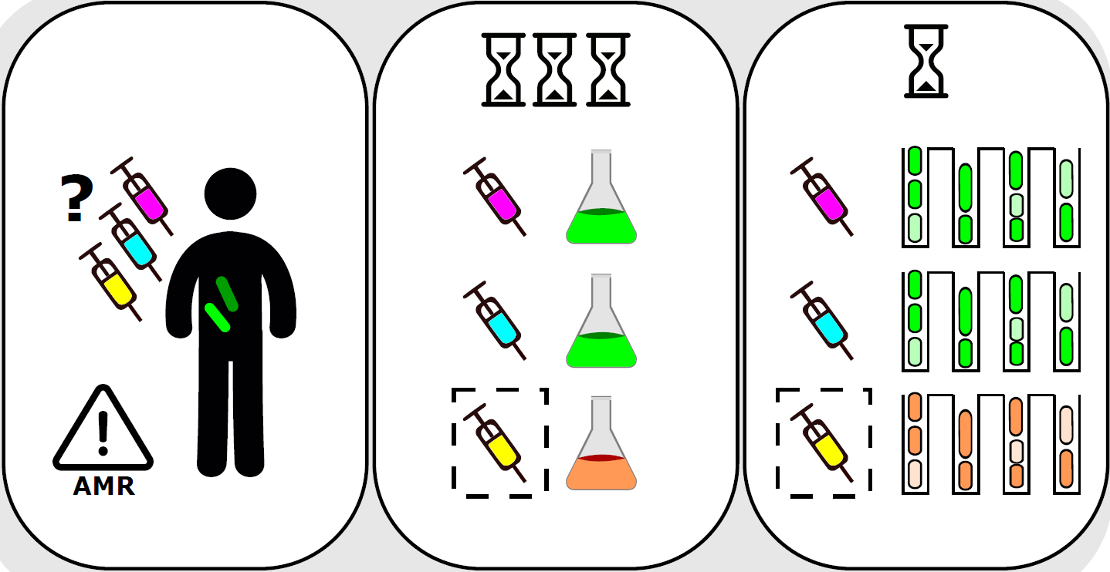
As antibiotic resistance is spreading, we need faster diagnostic tests to guide correct antibiotic prescription
A recent report estimates that by 2050, as many as ten million people could die each year as a result of antimicrobial resistance (AMR) [1]. One way to provide correct treatment and slow down the development of antibiotic resistance is to assay the susceptibility profile of the infecting bacteria before treatment is to enable personalized prescriptions in high-resistance environments and to limit the use of broad-spectrum drugs.
The limitations of conventional phenotypic ASTs (disk diffusion agar dilution or broth microdilution) are that they require bacterial growth for extended periods in the presence and absence of antibiotics to see an effect. However, for certain types of bacterial infections, even a delay of 6 h before treatment is initiated can have severe consequences [2].
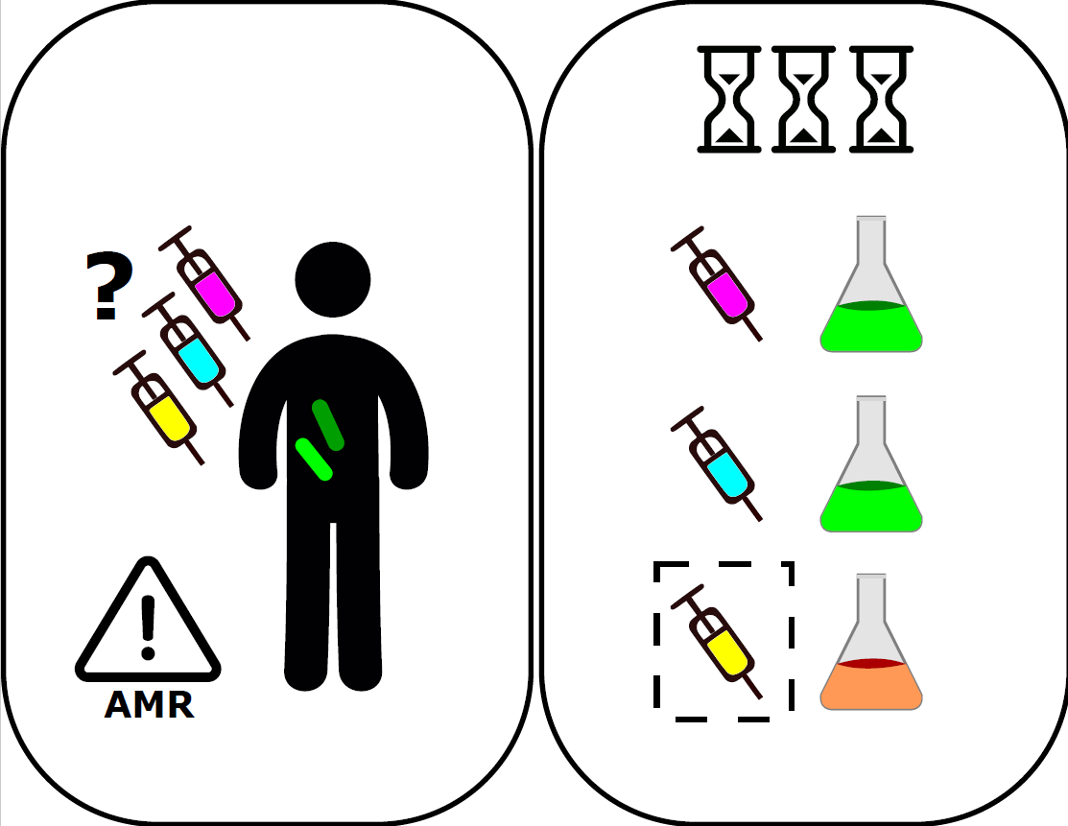
Figure 1: Antibiotic susceptibility tests in liquid cultures are reliable for detecting resistance and determining the antibiotic that prevents bacterial growth. These methods are usually very slow [Ilaria Iuliani]
Antibiotic susceptibility testing using direct single-cell imaging in microfluidic devices
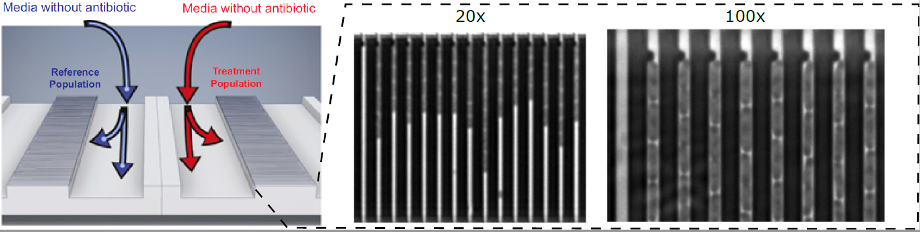
The device presented in this work has two rows of cell traps. In each row, there are 2,000 cell traps of dimension that can be loaded with bacterial cells from the front channel. A constriction at the end of each trap prevents cells from passing to the back channel, while allowing the media to flow around the cells.
The sample is loaded on the microfluidic chip and the bacteria present in the sample are caught in the bacteria-sized traps.
Trapping of bacteria is monitored, and the loading time gives an estimate of the bacterial density in the sample.
The reference population row receives the antibiotic-free medium, and the treatment population row receives the media with the antibiotic to be tested.
Bacterial growth is monitored in each trap, both with and without candidate antibiotic.
The average growth rates are calculated in real time, and bacteria are considered susceptible if its growth is duly inhibited.
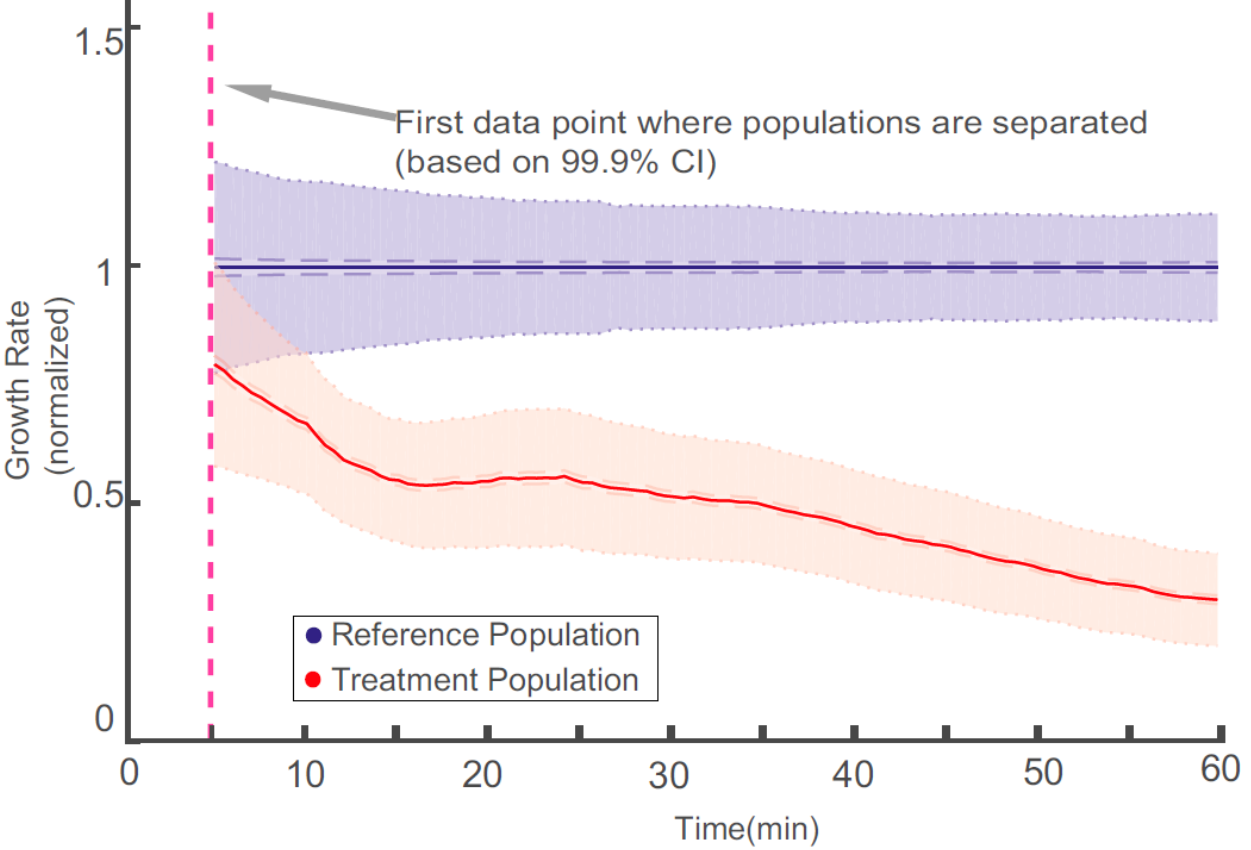
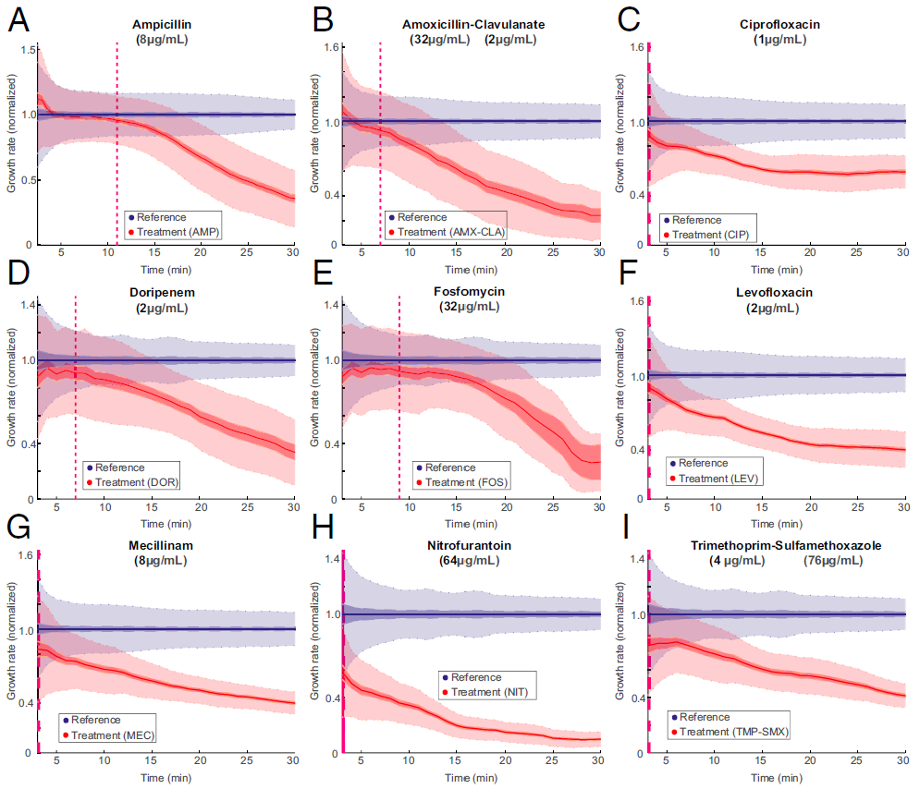
Figure 3: The overlay of the two populations normalized growth rate distributions. The time of separation of the treatment population from the reference population based on 99.9% CIs occurs before the dashed magenta line, which indicates the first time point when growth rates are estimated. [https://doi.org/10.1073/pnas.1708558114, PNAS].
An example: urinary tract infection by uropathogenic Escherichia coli
Nowadays, urinary tract infections (UTI) are very common, with 100 million cases of UTI per year worldwide and a high frequency of resistance to primary antibiotics.
For these infections, a fast AST could improve medical practice by making it possible to prescribe an antibiotic to which the infecting bacteria are susceptible before the patient leaves the primary care unit.
The total time for antibiotic susceptibility testing, from loading of sample to diagnostic readout, is less than 30 min, which allows the development of a point-of-care test that can guide correct treatment of urinary tract infection.
Although in this work they focused on bacterial species and antibiotics related to UTIs, the same principles would work for sepsis, mastitis, or meningitis.
Susceptibility testing and species identification for mixed samples
In the very recent paper, Elf group shows how to use this tool for combined phenotypic AST and species identification from mixed samples [3].
Current rapid phenotypic AST methods do not include species identification (ID), leaving time-consuming plating or culturing as the only available option when ID is needed to make the sensitivity call.
To meet this challenge, they used a deep learning method that can detect and track cells that come in different shapes and sizes allowing the analysis of growth-rates on multi-species samples, [4].
Subsequent genotyping was performed thanks to fluorescence in situ hybridization (FISH) using species-specific, fluorescent ssDNA probes.
Those early results seem promising, yet for this method to be clinically relevant, it will need to be calibrated and tested on many more clinical isolates as well as on real patient samples.
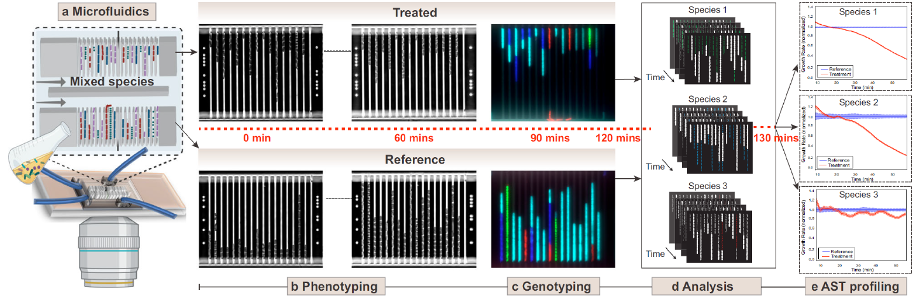
Figure 5: Schematic representation of the AST workflow with timeline for the mixed species loaded on the chip [ https://doi.org/10.1038/s41467-022-33659-1 ]
Why did I like this tool?
During my PhD, I have used a microfluidics device very similar to the one presented in this work to study the replication-division control in E.coli [5].
I found very amazing how J. Elf and his collaborators were able to use this relatively simple microfluidic device to very rapidly determine antibiotic susceptibility in dilute bacterial samples.
The chip design has been patented and the fast antibiotic susceptibility test has been developed into a product by the Astrego
company.
References:
[1] O’neill. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Review on antimicrobial resistance. https://amr-review.org/ (2014).
[2] Bernhard, M., Lichtenstern, C., Eckmann, C. & Weigand, M. A. The early antibiotic therapy in septic patients–milestone or sticking point? Crit. Care 18, 671 (2014). https://doi.org/10.1186/s13054-014-0671-1
[3]Kandavalli, V., Karempudi, P., Larsson, J. et al. Rapid antibiotic susceptibility testing and species identification for mixed samples. Nat Commun 13, 6215 (2022). https://doi.org/10.1038/s41467-022-33659-1
[4] O’Connor, O. M., Alnahhas, R. N., Lugagne, J.-B. & Dunlop, M. J. DeLTA 2.0: A deep learning pipeline for quantifying single-cell spatial and temporal dynamics. PLoS Comput. Biol. 18, e1009797 (2022). https://doi.org/10.1371/journal.pcbi.1009797
[5] https://www.elveflow.com/microfluidic-applications/microfluidic-cell-culture/how-to-study-bacteria-by-microfluidics/
MORE LIFE SCIENCE TOOLS TO FOLLOW
You want the latest on life science tools to follow, test and more?